2. Regression-based mapping of forest aboveground biomass
Authors: Sam Cooper, Akpona Okujeni, Patrick Hostert, Benjamin Jakimow, Andreas Janz, Fabian Thiel, & Sebastian van der Linden
Contributors: Klara Busse, Clemens Jaenicke
Publication date: 03/07/2020
Latest update: 23/01/2024
2.1. Introduction
2.1.1. 1. Background

This tutorial is featured on the HYPERedu online learning platform, an educational initiative under the EnMAP mission hosted on EO College. HYPERedu offers annotated slide collections and hands-on tutorials utilizing the open-source EnMAP-Box software, covering fundamental principles, methods, and applications of imaging spectroscopy.
Slide collections with annotations for the tutorial Regression-based mapping of forest aboveground biomass and a software description unit for the EnMAP-Box can be found here:
2.1.2. 2. Content
Forest aboveground biomass (AGB) is a measure of the living and dead plant material in a given area. As such, it is often used for forest management, assessing fire potential, and is an important metric used in modelling carbon and nutrient cycles. AGB can be directly measured at a plot level by harvesting and weighing vegetation, but this is both an expensive and highly invasive process. Through the use of statistical modelling and remotely sensed imagery, AGB can be mapped across broad, spatially continuous areas using only a small number of directly measured reference plots. This tutorial focuses on regression-based modeling of forest AGB using the EnMAP-Box. A hyperspectral image mosaic from the EnMAP sensor (here simulated from AVIRIS imagery) and a corresponding vector dataset containing plot-based AGB references are used for this tutorial. The aim is to provide an introduction into the functionality of the EnMAP-Box, as well as hands-on training for implementing regression-based mapping.
2.1.3. 3. Requirements
This tutorial is designed for EnMAP-Box 3, version 3.13.0 or higher. Minor changes may be present in subsequent versions, such as modified menu labels or added parameter options.
2.1.4. 4. Further reading
We recommend [1] and [2] for a comprehensive overview of imaging spectroscopy of terrestrial ecosystems, [3] for an overview of remote sensing of forest AGB and [4] for a companion study using the same data.
2.1.5. 5. Data
You can download the data for this exercise here:
The tutorial data contains a simulated hyperspectral EnMAP image, plot-based AGB references as well as a land cover map for a small study area located in Sonoma County, California, USA. The simulated EnMAP image is a subset extracted from the “2013 Simulated EnMAP Mosaics for the San Francisco Bay Area, USA” dataset [5]. AGB reference data was sampled from an existing LiDAR derived AGB map [6]. The land cover map was taken from the 2011 National Landcover Database (NLCD) [7].
Data type |
Filename |
Description |
|---|---|---|
Raster |
|
Simulated spaceborne hyperspectral data from the EnMAP sensor with a spatial resolution of 30m, 195 bands, and 1000x200 pixels (GeoTIFF |
Raster |
|
National Land Cover Database 30m classification for the study region (GeoTIFF |
Vector |
|
343 AGB reference points sampled from the existing LiDAR derived AGB map (GeoPackage |
Dubayah, R.O., A. Swatantran, W. Huang, L. Duncanson, H. Tang, K. Johnson, J.O. Dunne, and G.C. Hurtt. 2017. CMS: LiDAR-derived Biomass, Canopy Height and Cover, Sonoma County, California, 2013. ORNL DAAC, Oak Ridge, Tennessee, USA. https://doi.org/10.3334/ORNLDAAC/1523
Cooper, S.; Okujeni, A.; Jänicke, C.; Segl, K.; van der Linden, S.; Hostert, P. (2020): 2013 Simulated EnMAP Mosaics for the San Francisco Bay Area, USA. GFZ Data Services. https://doi.org/10.5880/enmap.2020.002
Multi-Resolution Land Characteristics Consortium (MRLC) (2018). National Land Cover Database 2011 (NLCD 2011). Multi-Resolution Land Characteristics Consortium (MRLC). https://data.nal.usda.gov/dataset/national-land-cover-database-2011-nlcd-2011. Accessed 2022-08-08.
2.2. Exercise A: Getting started with the EnMAP-Box
Description
This exercise introduces basic functionalities of the EnMAP-Box for this tutorial. You will get to know the graphical user interface and will learn how to load data, visualize raster and vector data, and use the basic navigation tools. Additionally, you will learn to work with multiple map views and how to visualize image spectra using Spectral Library Windows.
Duration: 30 min
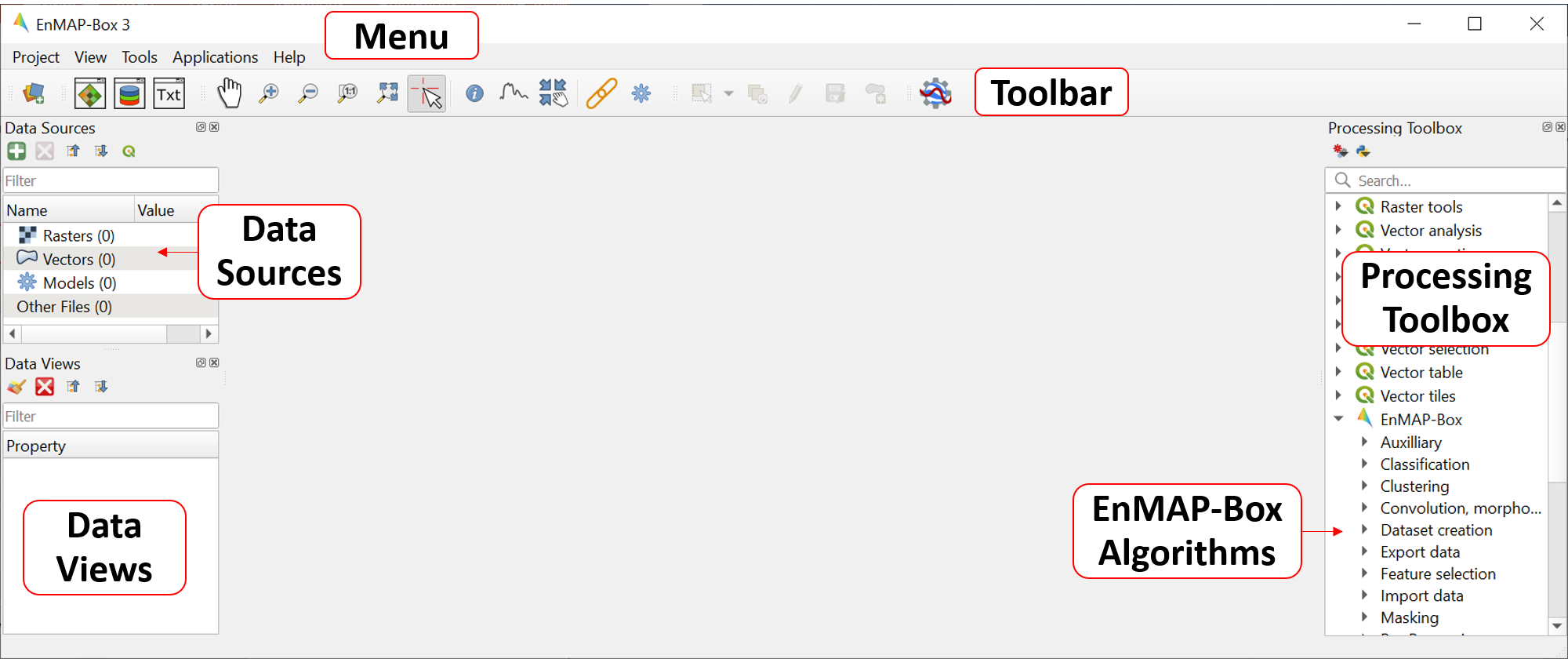
2.2.1. 1. Start the EnMAP-Box
Start QGIS and click the
 icon in the toolbar to open the EnMAP-Box. The GUI of the EnMAP-Box consists of
a Menu and a Toolbar as well as panels for Data Sources and Data Views.
icon in the toolbar to open the EnMAP-Box. The GUI of the EnMAP-Box consists of
a Menu and a Toolbar as well as panels for Data Sources and Data Views.The QGIS Processing Toolbox including the EnMAP-Box algorithms can be optionally added to the GUI by clicking on View in the Menu and by checking the Processing Toolbox from the Panel list.
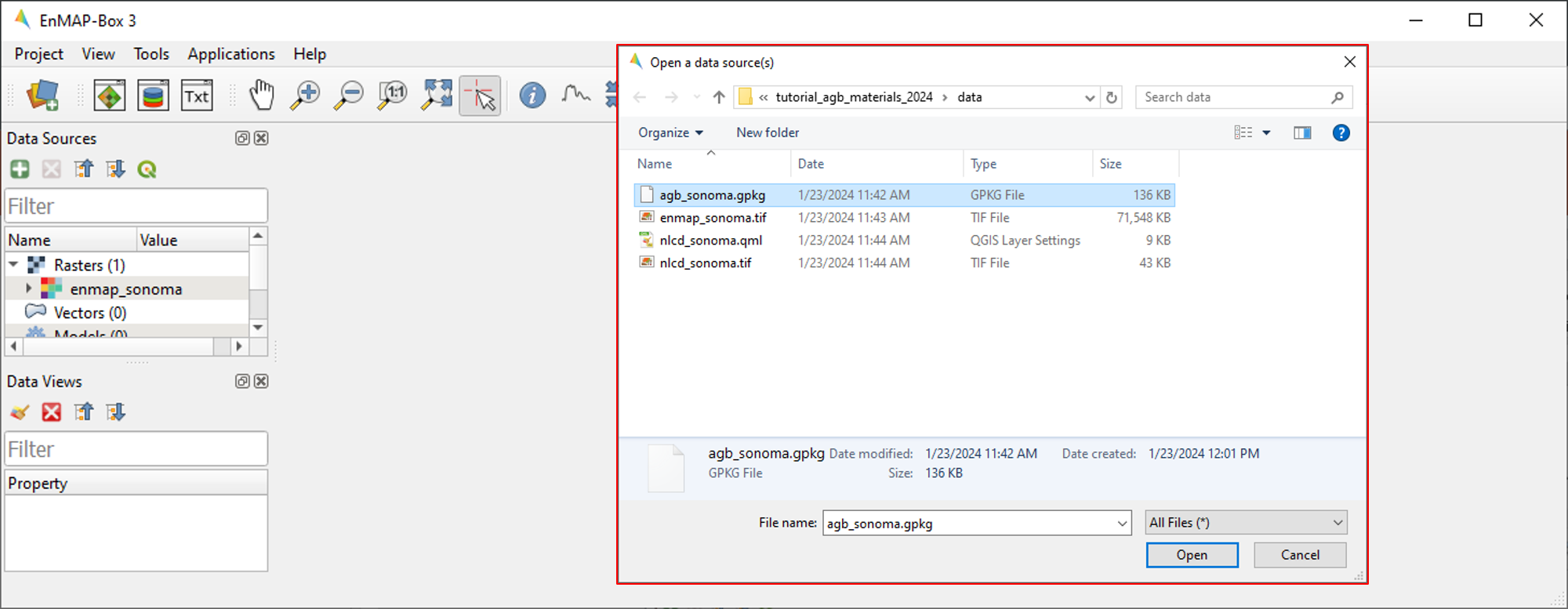
2.2.2. 2. Load data00
To load new datasets into the EnMAP-Box, click the
icon and navigate to the file directory containing your data. Select
agb_sonoma.gpkgfrom the Open data source dialogue and select Open.Alternatively, the EnMAP-Box offers simple drag & drop capabilities to load data from an external file manager (e.g. Windows File Explorer). Load
enmap_sonoma.tifby dragging and dropping the file from your file manager into the Data Sources panel.All data currently open in the EnMAP-Box will appear in the Data Sources panel.
2.2.3. 3. Visualize raster data
The EnMAP-Box offers Map Views (Map #) for visualizing raster and vector data. Click the
icon to open a new Map View and drag
enmap_sonoma.tiffrom the Data Sources panel into Map #1.In addition to a new Map View opening, a corresponding Data View entry is created in the Data Views panel which shows all data currently loaded in a given Map View.
The
enmap_sonoma.tifimage will be displayed as true color RGB composite. True color rendering is based on predefined RGB band combinations (R: 658 nm, G: 569 nm, B: 479 nm) stored in the QGIS Style Fileenmap_sonoma.qml.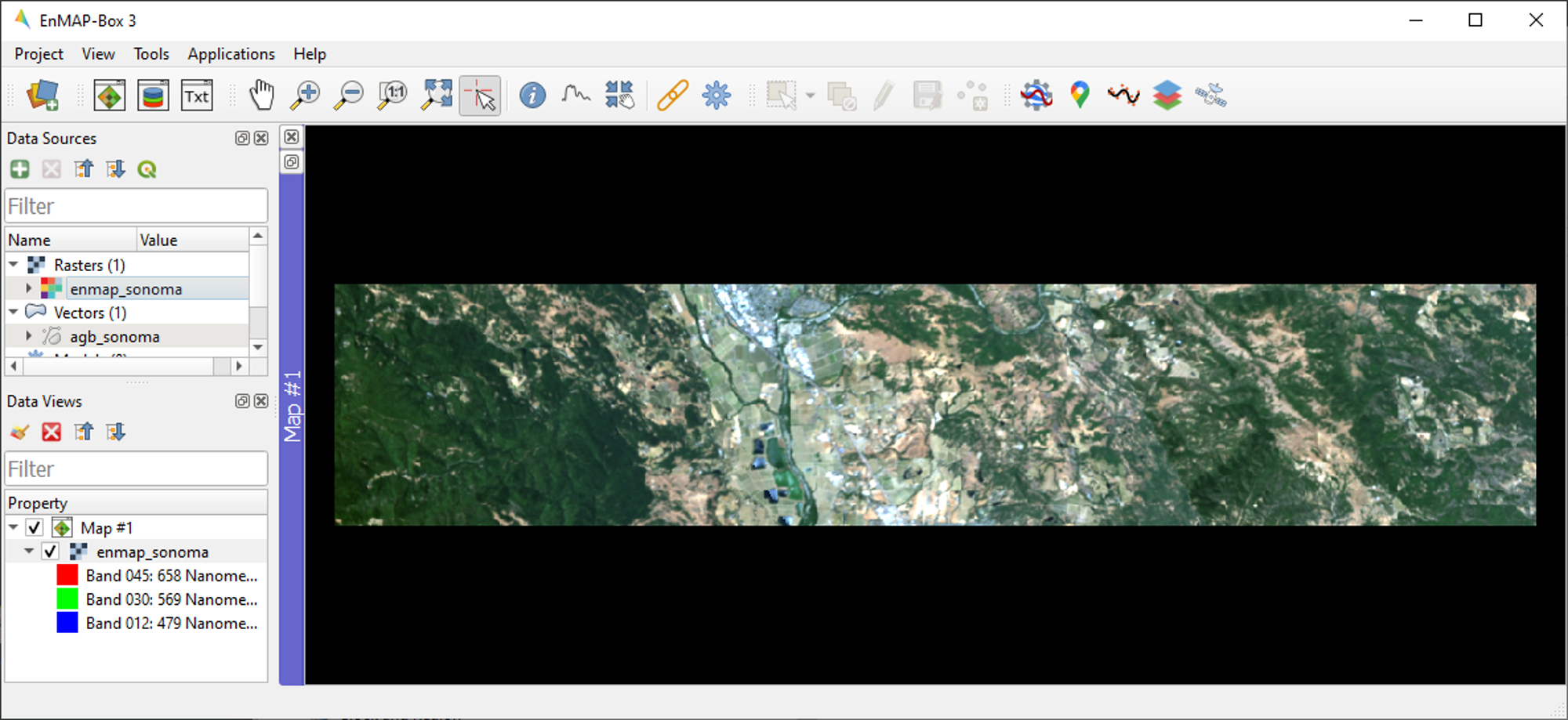
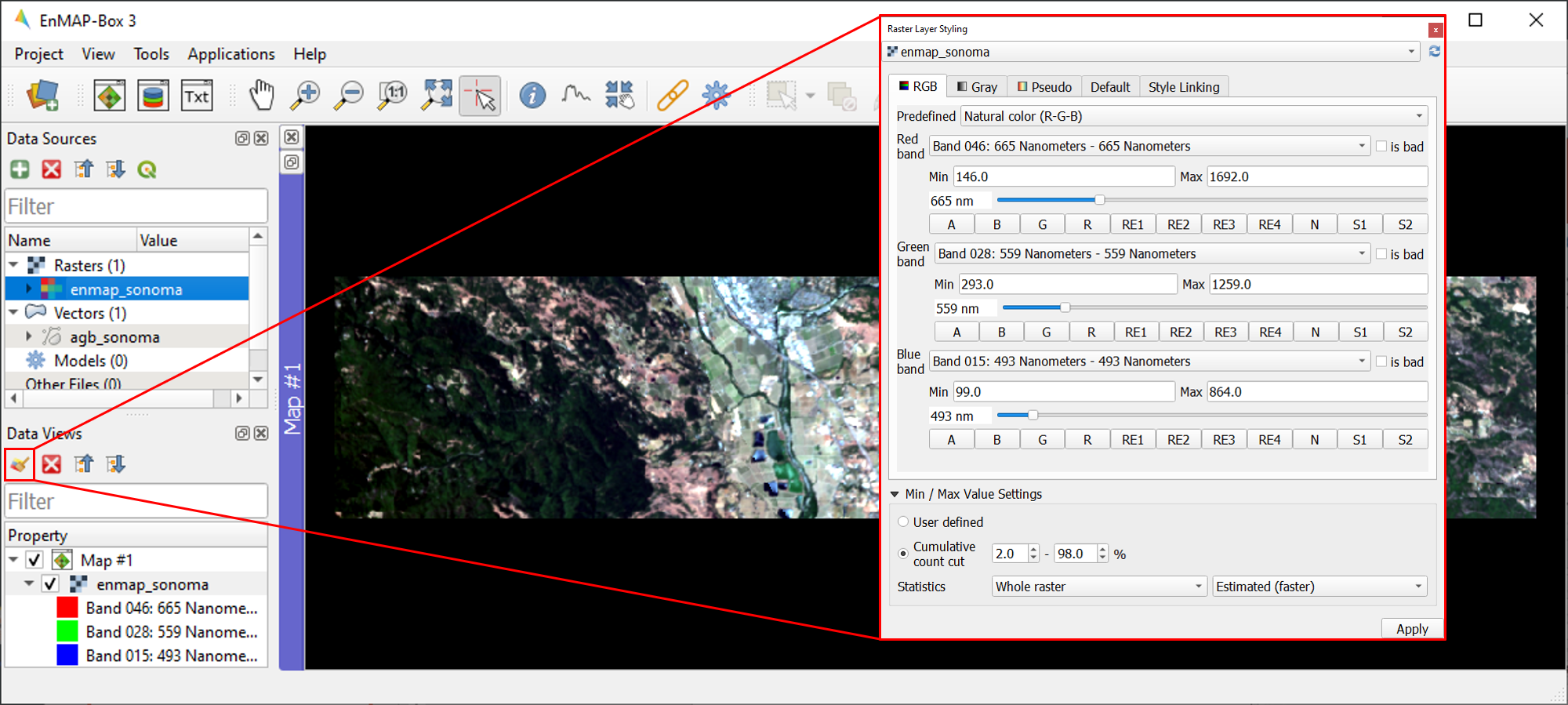
Raster Layer Styling is a versatile tool for defining or modifying the RGB representation of raster images, either manually or through pre-defined settings. To change the RGB representation of
enmap_sonoma.tif, open the Raster Layer Styling panel by clicking theicon in the Data Views panel.
Choose
enmap_sonoma.tiffrom the dropdown menu and the RGB tab as render type.You can now choose between various pre-defined RGB representations from the dropdown menu (note: rasters needs to have wavelength information) or manually specify your RGB band combination.
You may further use the functionalities offered for contrast enhancement (e.g. Cumulative count cut: 2-98%).
2.2.5. 5. Multiple map views
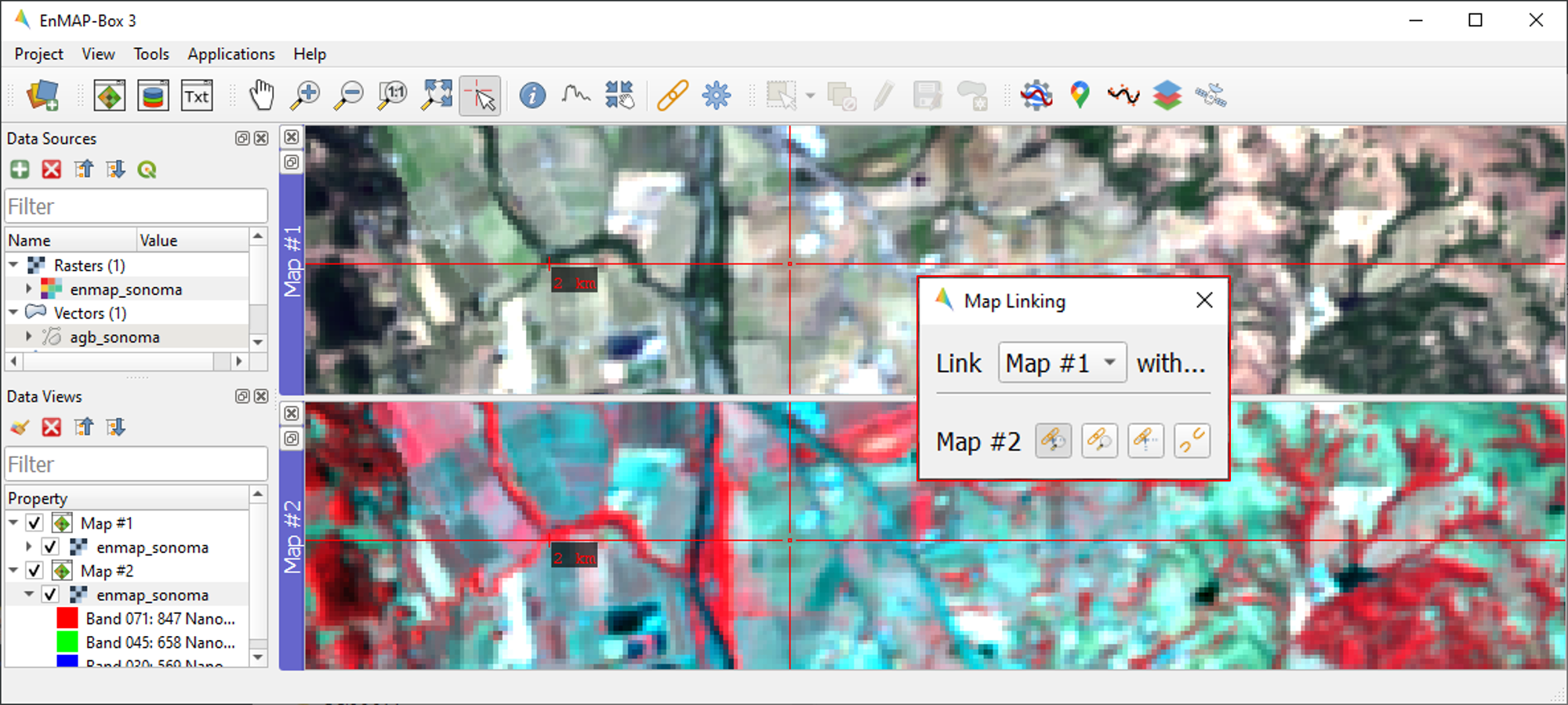
The EnMAP-Box enables users to work with multiple Map Views, which can be flexibly organized and geospatially linked.
Open a new Map View (Map #2) by clicking the
icon. A new Data view appears corresponding to the newly added Map View.
Display
enmap_sonoma.tifas an RGB composite of your choice in Map #2.
Tip
When loading a raster image to a map view, you may also right click the filename in the Data Sources panel and select either Open in existing map or Open in new map. If the raster image has wavelength information associated with it, you may also select a predefined composite from the context menu.
To establish geospatial linking, click the
icon to open the Map Linking window. Choose the
Link Map Scale and Center option, and close the dialog.
Tip
Map Windows can be re-arranged by clicking on the blue Map title bar (Map #) and dragging it to the desired position.
A transparent blue rectangle will appear indicating the docking position once you release the mouse button.
You may also undock map views from the EnMAP-Box window by selecting  from the blue Map title bar.
To re-dock a Map View, click and drag the blue Map title bar to an open Map View already docked in the EnMAP-Box window.
from the blue Map title bar.
To re-dock a Map View, click and drag the blue Map title bar to an open Map View already docked in the EnMAP-Box window.
2.2.6. 6. Visualize vector data
Close Map #2 from the previous step.
Load
agb_sonoma.gpkgto Map #1.To change the order of stacked layers, go to the Data Views panel and drag one layer on top or below another one. Arrange the layer stack so that
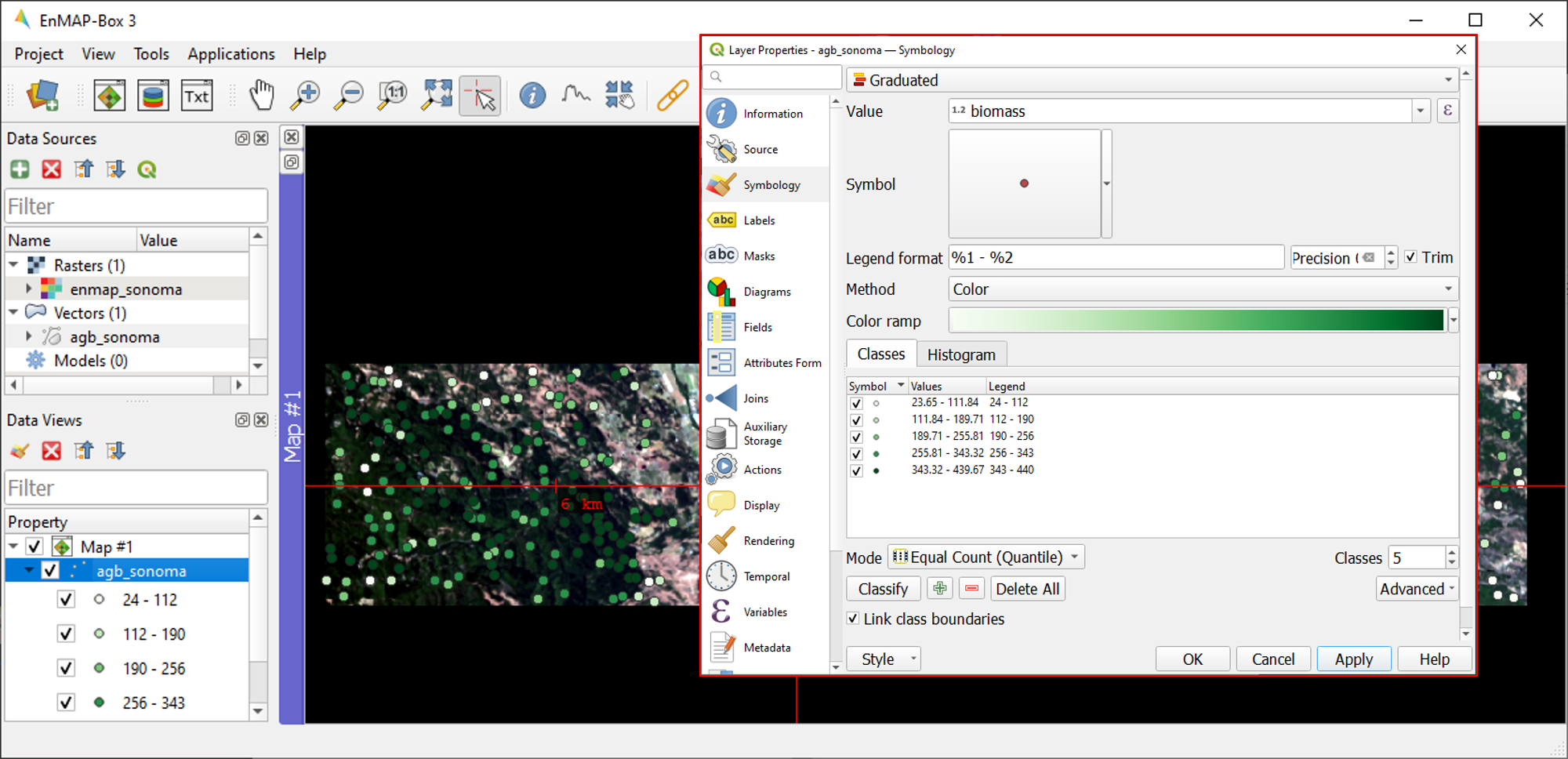
agb_sonoma.gpkgis displayed on top ofenmap_sonoma.tif.By default, vector files are displayed with a single uniform symbol. To change this symbology, right click on
agb_sonoma.gpkgin the Data Views panel, select Layer Properties and navigate to Symbology in the Layer Properties window. You can now change the symbology in accordance to the QGIS functionality.Select Graduated from the dropdown menu, and select
biomassin Value andColorin Method.Set the Color ramp to run from white to green.
Press Classify and then OK to display the biomass values associated with each point.
2.2.7. 7. Extract & visualize image spectra
The EnMAP-Box offers Spectral Library Windows (SpectralLibrary #) for visualizing spectra and handling their metadata.
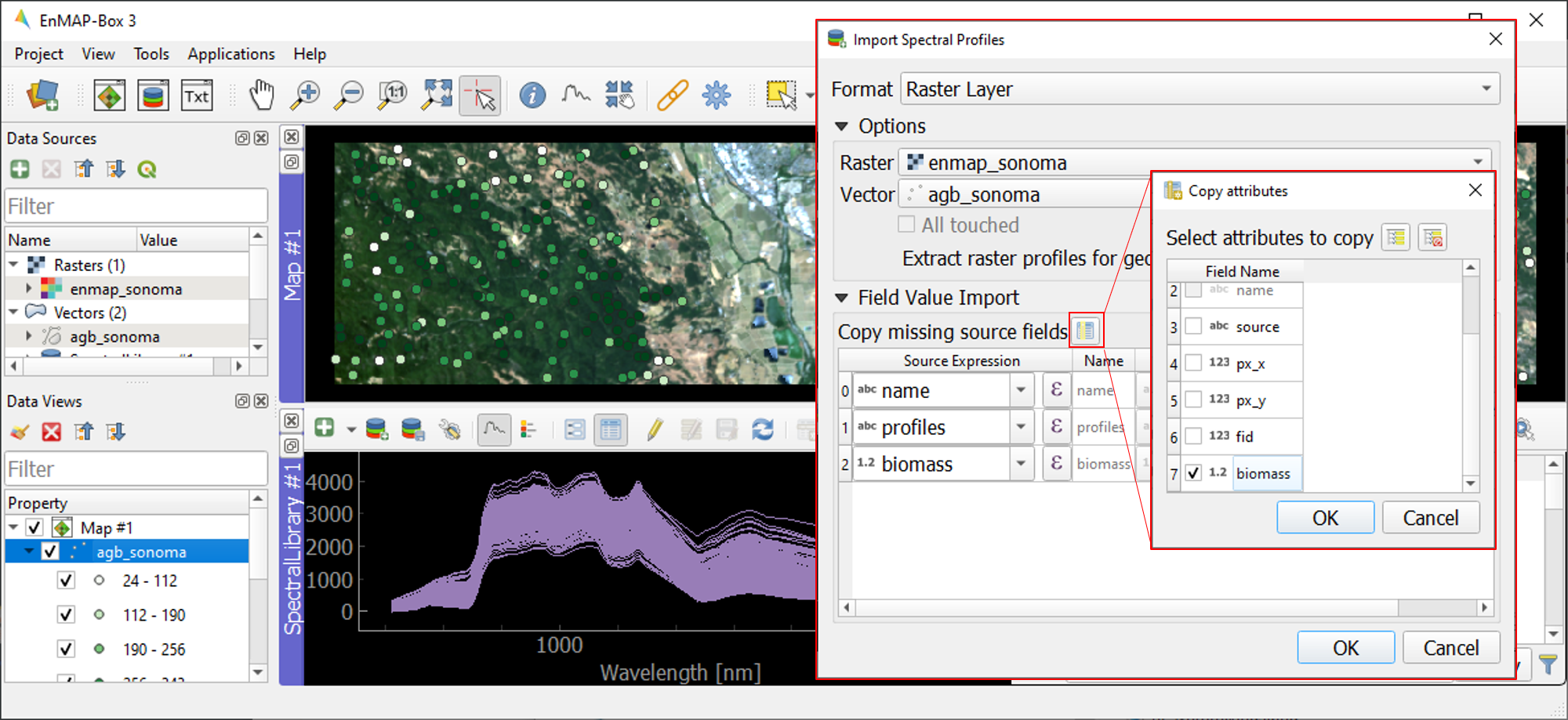
This tool may also be used to extract and visualize spectra which are spatially associated with vector data open in the EnMAP-Box, i.e., the AGB reference points. To do this, open a new Spectral Library window by selecting the
icon on the toolbar.
Next, import spectral profiles from other sources by clicking at the
icon in the SpectralLibrary #1 menu. Specify the following settings:
Terminate the Import Spectral Profile dialogue with OK. A spectral library is automatically built based on the geographic location of each point in the vector file. The associated attribute information is displayed in the table on the right.
In Exercise B, you will learn how to create regression models based on the illustrated spectra and related AGB quantities to predict AGB across the whole image.
Learning Activities
A1: What land cover types are present in the imagery? How are the AGB reference plots distributed throughout the scene?
A2: What different information can you see when switching from a true color composite to a NIR false color composite?
2.3. Exercise B: Regression based mapping of AGB
Description
One of the strengths of remote sensing comes from its ability to take high-quality plot measurements of a variable of interest and building statistical models with which wall to wall maps of this variable can be created. One of the most common ways of doing this is to create regression models based on the optical properties of the training data and applying it to large scale imagery. This exercise …
Introduces a regression-based mapping approach for taking plot measurements of AGB and generating spatial AGB estimates using an input raster of hyperspectral imagery.
Demonstrates the Regression Dataset Manager and the Regression Workflow applications of the EnMAP-Box.
Duration: 20 min
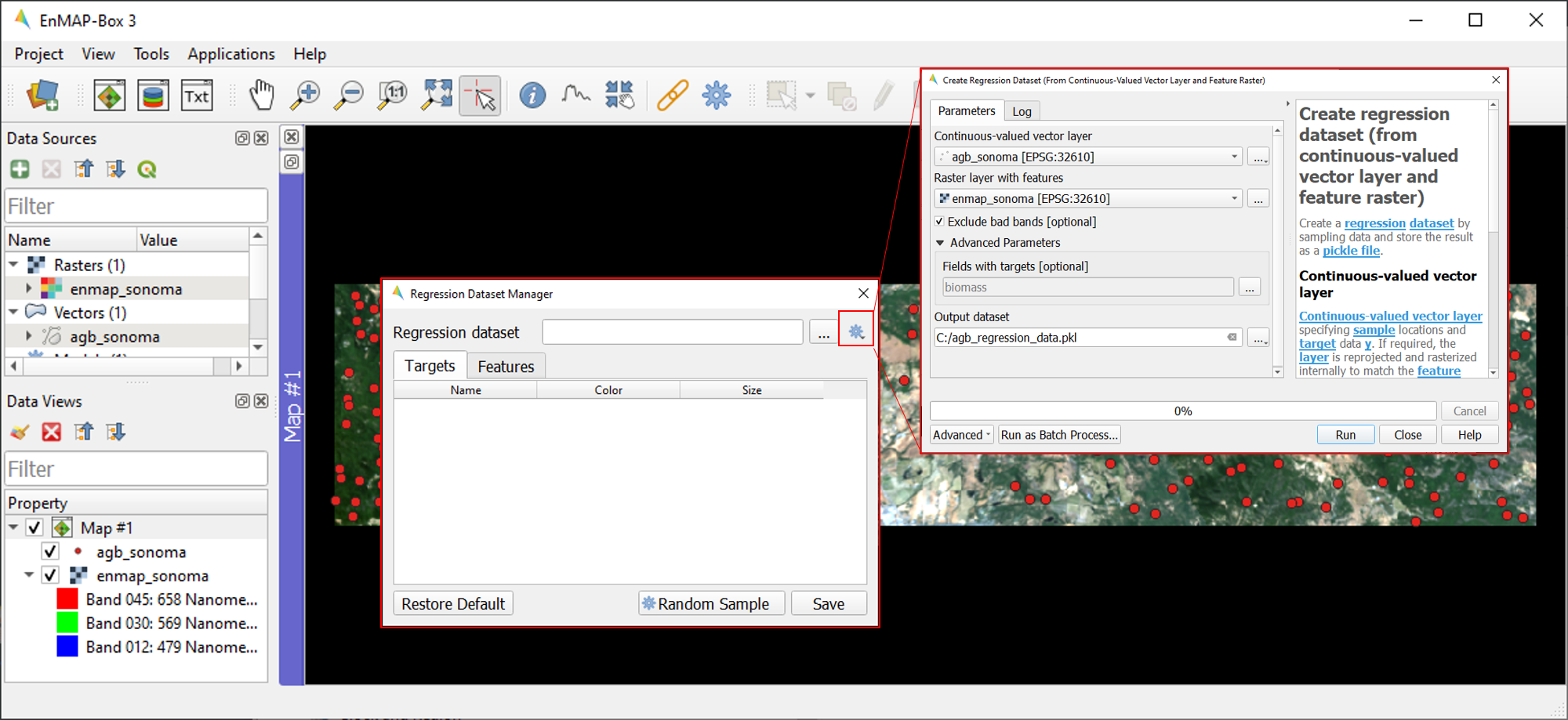
2.3.1. 1. Use the Regression Dataset Manager for data preparation
The Regression Dataset Manager offers different options to prepare data for the Regression workflow application. In the context of this tutorial, you will create a Regression Dataset from a raster and a vector layer containing the spectral features (independent variable) and the target variable (dependent variable), respectively. The regression dataset will be stored as pickle file (
.pkl).Open
enmap_sonoma.tifandagb_sonoma.gpkgin a single Map Window. Close all other opened Map and Spectral Library Windows.Navigate to Applications in the Menu, choose Regression, and then Regression Dataset Manager.
To create the Regression Dataset from a raster and a vector layer, click on the
icon and select
Create regression dataset (from continuous-valued vector layer and feature raster). A new widget will be opened. Run the dialog with the following inputs:
mIconPolygonLayer
Continuous-valued vector layer: select
agb_sonoma.gpkgRaster layer with features: select
enmap_sonoma.tifFields with targets: select attribute
biomassOutput dataset: select and define an output path and file name (e.g.
agb_regression_data.pkl).
After running the dialog,
agb_regression_data.pklwill be opened under Models in the Data Sources panel. Close the Regression Dataset Manager.
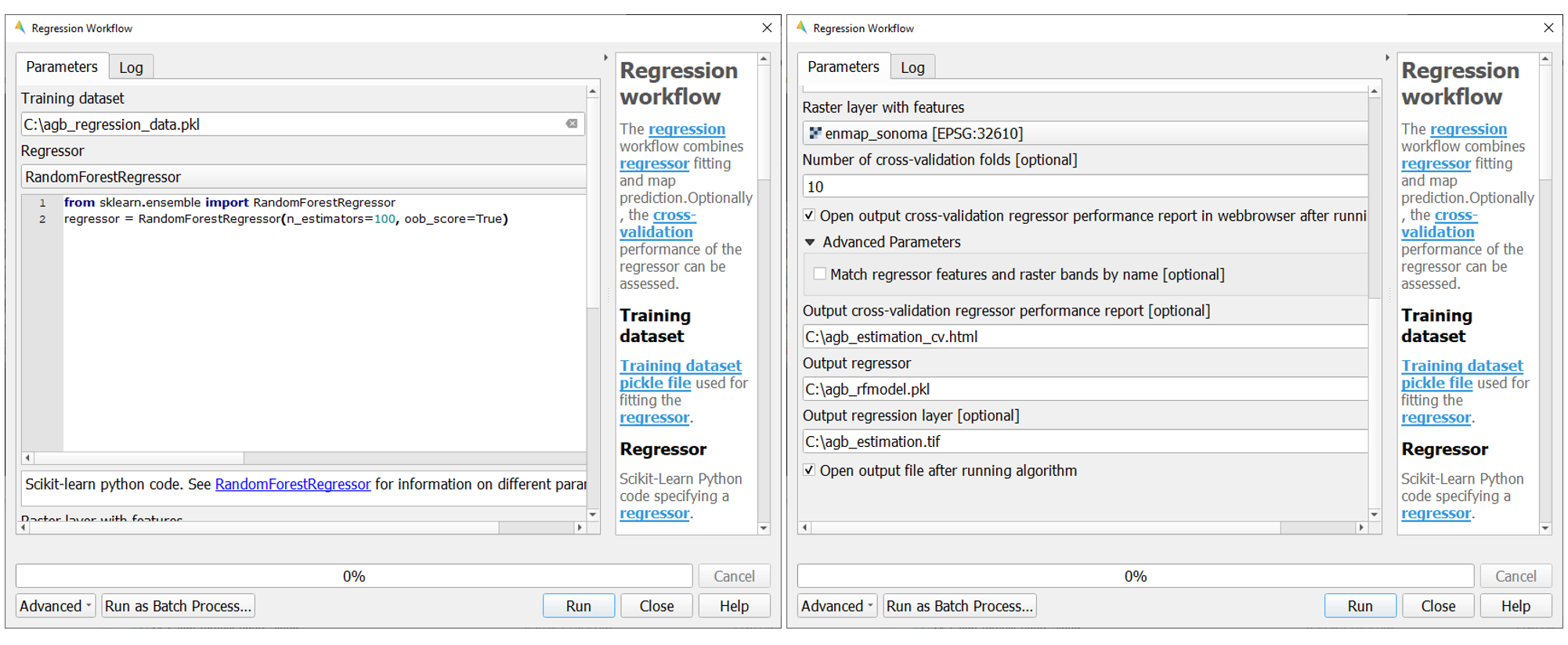
2.3.2. 2. Use the Regression Workflow for estimating AGB
The Regression Workflow application offers several state-of-the-art regression algorithms from the scikit-learn library (see https://scikit-learn.org/stable/index.html) for predicting continuous variables. The application further includes an optional cross-validation for assessing model performances.
Click on Applications in the Menu, select Regression, and then Regression workflow to open the regression application.
Choose
agb_regression_data.pklas Training dataset.Select
RandomForestRegressor(default, due to the low processing time) as Regressor, and use the default model parameters. Note that the different algorithms provided lead to varying accuracies and processing times. Refer to the scikit-learn documentation for more information.Raster layer with features specifies the raster image to which the regression model will be applied. Select
enmap_sonoma.tif.To make use of a cross-validation, set the Number of cross-validation folds’ to ``10` (default) and leave the Open output cross-validation regressor… option checked. Specify output path and file name (e.g.
agb_estimation_cv.html) under Output cross-validation regressor performance report to save the report in your working directory.The regression model can be optionally saved, e.g. for applying the model again to a dataset. Specify output path and file name (e.g.
agb_rfmodel.pkl) under Output regressor to save the result in your working directory.Specify output path and file name (e.g.
agb_estimation.tif) under Output regressor layer to save the result in your working directory.Click run to start the Regression Workflow.
Tip
All processing options of the Regression Workflow that are labeled as [optional] can be disregarded by
setting the Output to Skip Output.
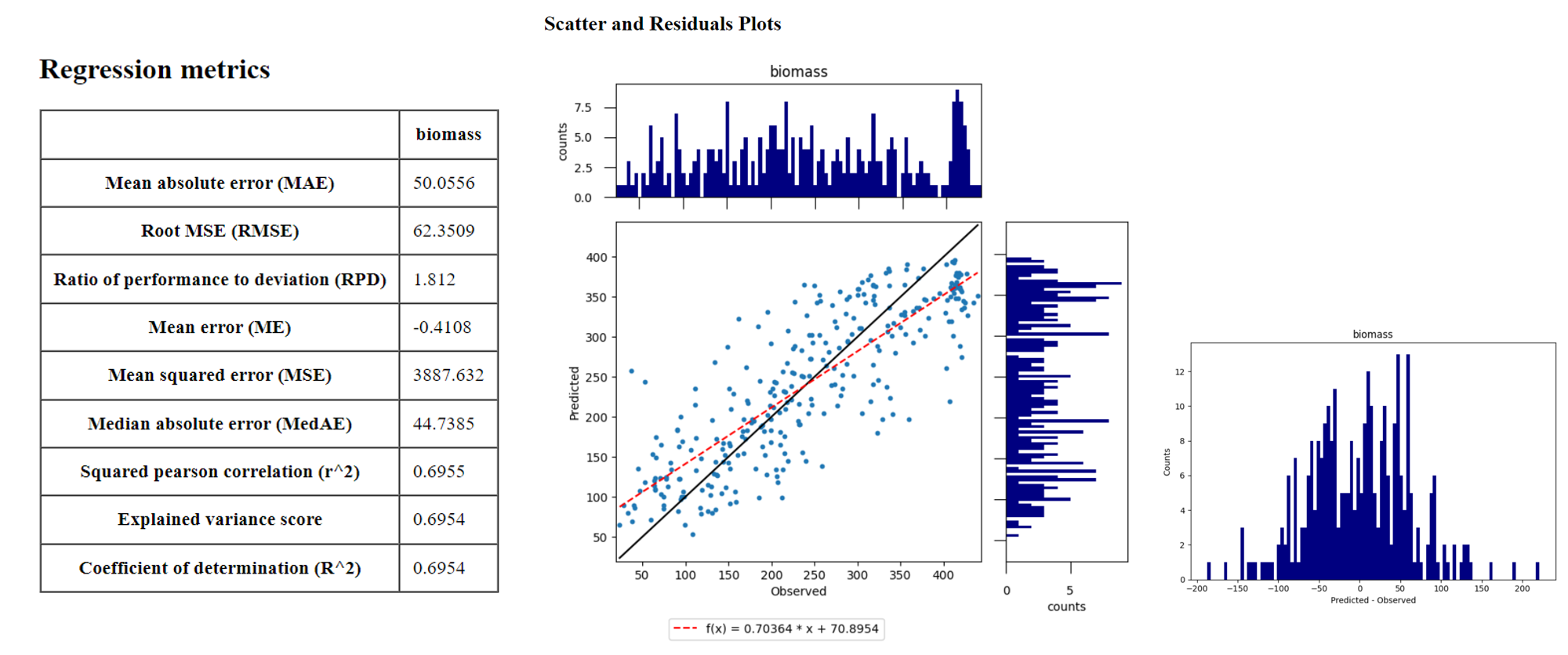
2.3.3. 3. Assess the model performance for AGB estimation
After running the Regression Workflow, the performance report with scatterplots and statistical measures will be opened in your default web browser.
Based on the 10-fold cross-validation, you can now access the performance of your model to predict AGB.
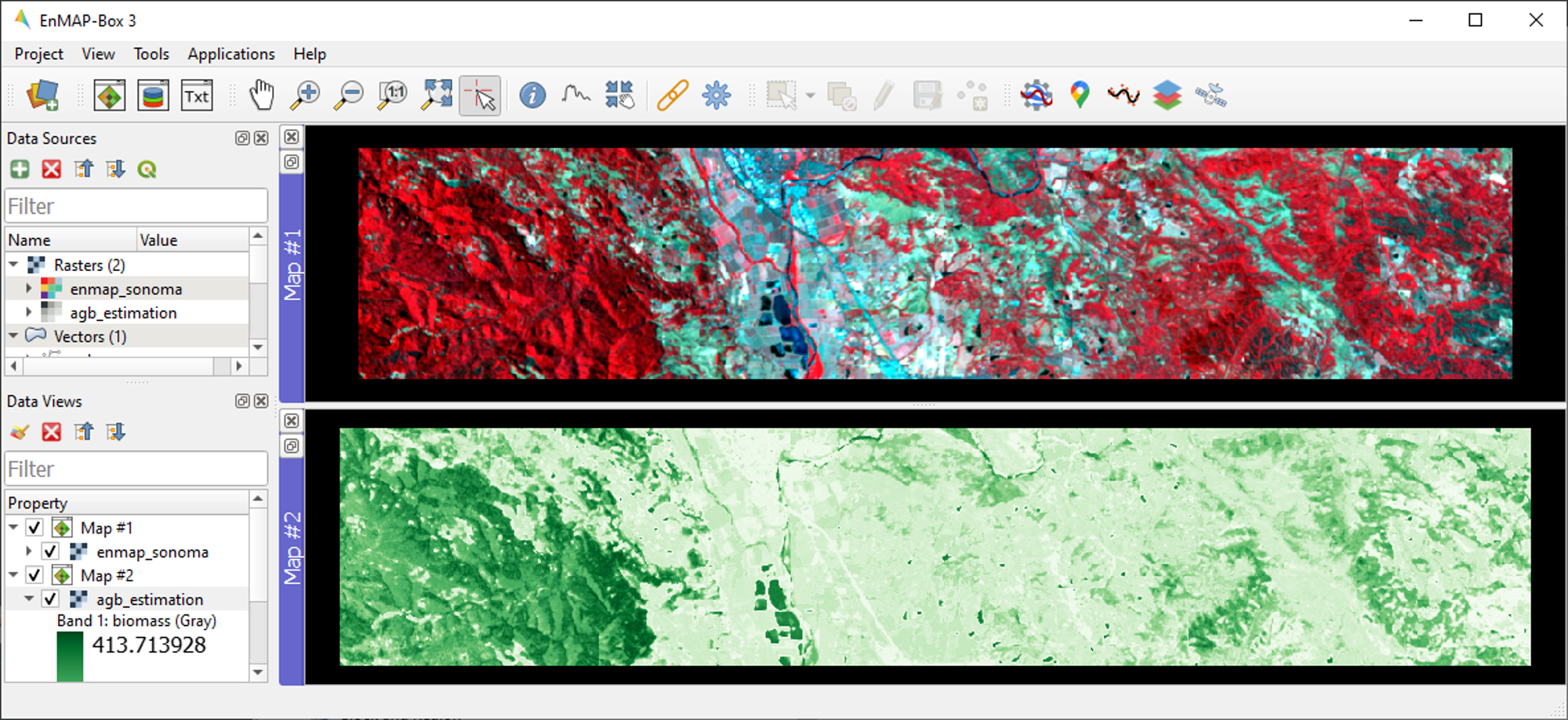
2.3.4. 4. Visualize AGB results
After running the Regression Workflow, all outputs will appear in the Data Sources panel.
Close all opened Map/SpectralLibrary Windows. Open
enmap_sonoma.tifas an RGB composite of your choice in Map #1.Open the
agb_estimation.tifin a new Map View and link to the Map #1. Use the Layer Properties to change the color ramp to white-green (Singleband pseudocolor).
2.3.5. Learning Activities
Learning Activities
B1: What general trends do you see in the biomass estimations? How do they compare to landscape features seen in the EnMAP imagery?
B2: Discuss the accuracy results, both in terms of the statistical measures, as well as the form of the scatterplot and histograms.
2.4. Exercise C: Compare AGB estimates with the NDVI
Description
In this exercise, you will learn how to use the Raster Math application to calculate a NDVI map and generate a forest mask based on the NLCD land cover map. Based on the forest area only, you will then assess the AGB prediction from Exercise B relative to the NDVI using the Scatter Plot Tool.
Duration: 30 min
2.4.1. 1. Introduction to Raster Math
Raster math is a powerful raster calculator inspired by the QGIS Raster calculator, the GDAL Raster calculator and ENVI Band Math. In addition to those tools, the EnMAP-Box Raster math calculator supports multi-band arrays, vector layer inputs, multi-line code fragments and metadata handling. In the following sections, you will utilize Raster Math to…
to calculate a NDVI map from two bands of our EnMAP imagery,
to generate a forest mask from the NLCD land cover map,
and to apply a forest mask to both the NDVI and AGB maps.
Close all opened Map/Spectral Library Windows. Display
enmap_sonoma.tif,nlcd_sonoma.tifandagb_estimation.tifin a single or in multiple Map Views.Open the ** Raster Math** application by going to Applications then selecting Raster math
Raster Math consists of several panels, including the :
Code: text editor to define a single-line expression or a multi-line code fragment for raster calculations on input datasets
Data sources: list of available datasets to conduct the calculations
2.4.2. 2. Calculate NDVI
The Normalized Difference Vegetation Index (NDVI) is a commonly used vegetation index that is correlated with both vegetation cover and AGB. The formula for NDVI is:
\[NDVI = \frac{NIR-Red}{NIR+Red}\]where NIR is the near-infrared band reflectance (~850nm) and Red is the red band reflectance (~660nm). We will now calculate NDVI from the EnMAP imagery using Raster Math.
In the Code Editor, define the Red and NIR bands in our EnMAP imagery. These correspond to bands 45 (658 nm) and 71 (847 nm), respectively:
red = enmap_sonoma@45 nir = enmap_sonoma@71
… then, define the NDVI formula:
ndvi = (nir - red) / (nir + red)
… and, finally, delete the temporary variables used for calculating the NDVI:
del red,nir
Specify output path and file name (e.g.
ndvi.tif) under Output raster layer to save the result in your working directory.Click run to start the calculation. A new raster dataset
ndvi.tifwill appear in the Data Sources panel.

2.4.3. 3. Create a forest mask
As the model was trained using AGB reference plots from forest areas, only limited inference can be made of the non-forest AGB estimates. We will therefore apply a forest mask to our AGB map as well as to the NDVI map. The forest mask will be generated based on the available NLCD land cover map.
Below are the NLCD classes and color legend represented in the raster data. We will consider any pixel to be forest which is labelled as Deciduous (41), Evergreen (42), or Mixed (43) forest according to the NLCD classification.
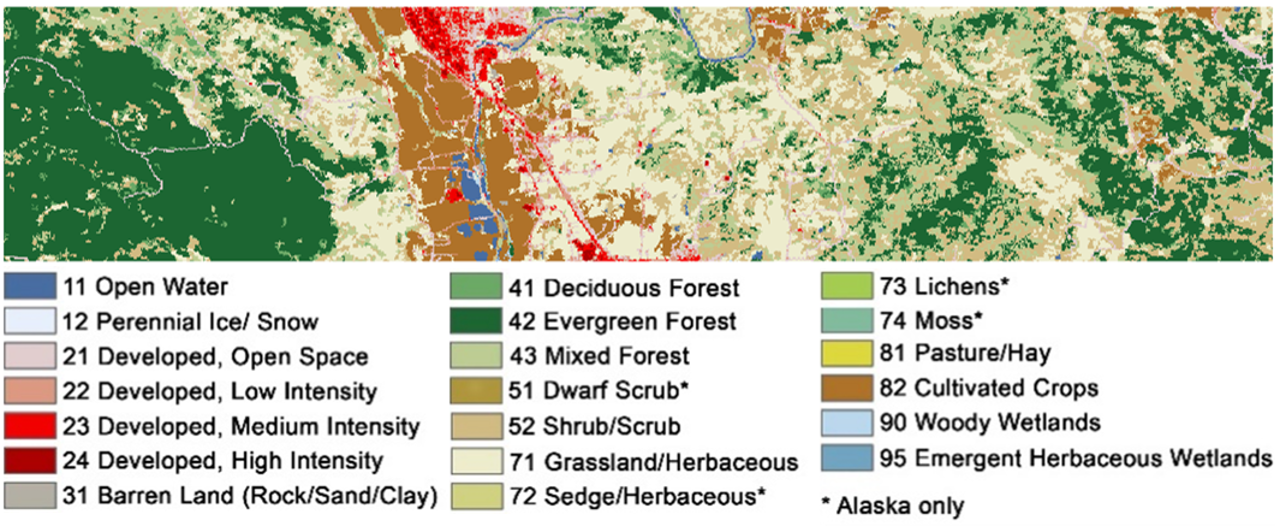
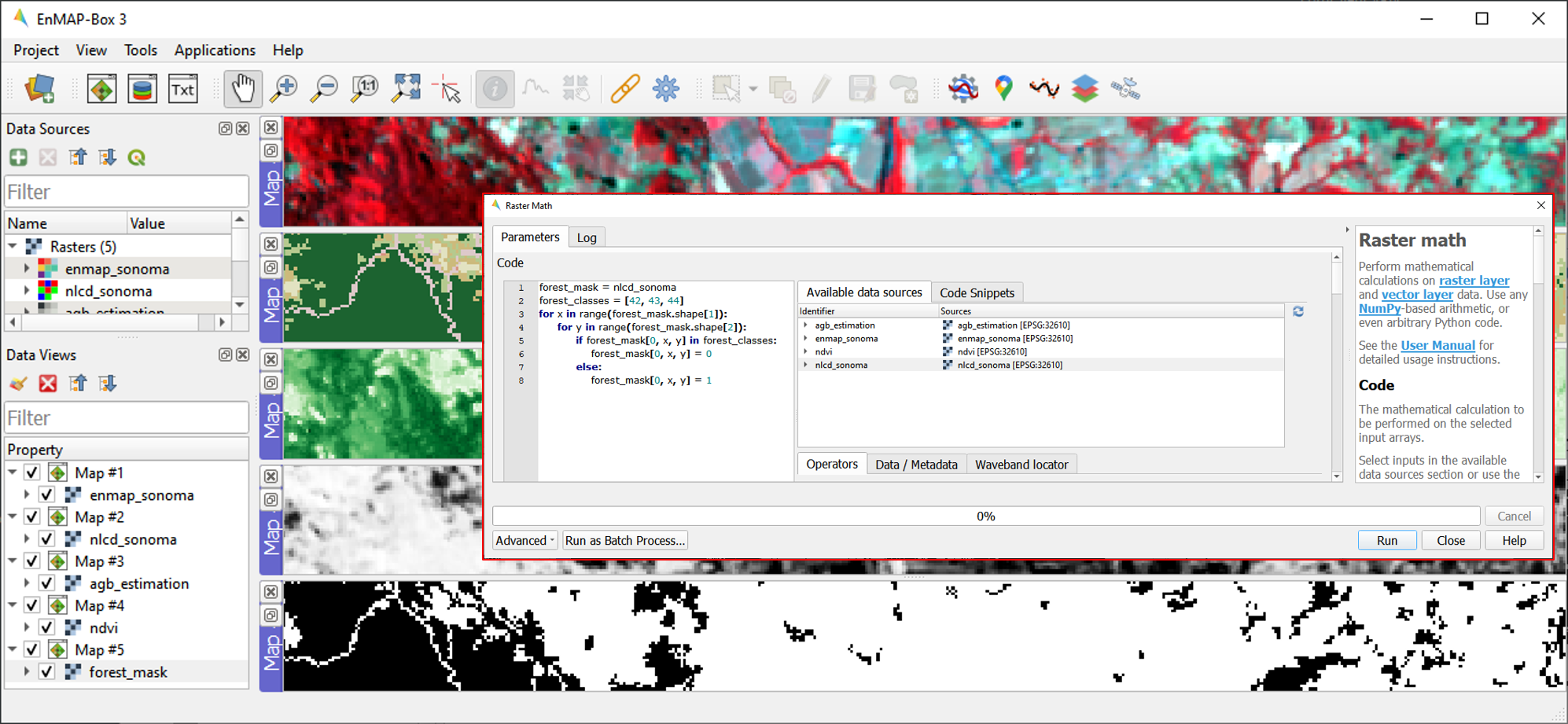
Open the Raster Math application and enter the following code into the code editor to calculate the forest masks:
forest_mask = nlcd_sonoma forest_classes = [42, 43, 44] for x in range(forest_mask.shape[1]): for y in range(forest_mask.shape[2]): if forest_mask[0, x, y] in forest_classes: forest_mask[0, x, y] = 0 else: forest_mask[0, x, y] = 1
Line by line, this
Copies the NLCD information to a new object we will manipulate to create the mask
Creates a list of classes which we consider forest
Loops through the x dimension of the raster.
Loops through the y dimension of the raster.
Check if the element at the current x and y position is in the forest_classes list
If it is, set that value to 0
Else
Set that value to 1
Specify output path and file name (e.g.
forest_mask.tif) under Output raster layer to save the result in your working directory.Click run to start the calculation. A new raster dataset
forest_mask.tifwill appear in the Data Sources panel. The resulting mask now has a value of 0 for forested pixels, and 1 for non-forested pixels.
2.4.4. 4. Apply the forest mask to the AGB estimates
Open the Raster Math application and enter the following code into the code editor to apply the forest mask to the AGB and NDVI images.:
mask = forest_mask.astype(bool)
agb_masked = agb_estimation
agb_masked[mask] = -99
agb_masked.setNoDataValue(-99)
del mask
Line by line, this script:
Sets the mask to a Boolean data type (i.e. True/False). The mask file contains binary values where 0 indicates forest (i.e. non-masked pixels) and 1 indicates non-forest (i.e. pixels to be ignored). In Python, 1 also represents True while 0 represents False, and by setting the datatype to
bool, we explicitly tell Python to treat these values in this manner.Copies the AGB values to a new array.
Steps through each value in the new array and sets the value to -99 if the mask value is True. In numpy array speak, this line can therefore read: “For each value in
agb_masked, if the corresponding value inforest_maskis True (i.e. non-forest), then set that value to -99”. If the mask value is False (i.e. forested), nothing will happen, and the biomass value will remain in the array.Sets the no data value for the masked array to -99. This helps the EnMAP-Box to automatically display the data correctly, and since it is not a realistic value for both AGB and NDVI, we can safely ignore it.
Deletes the temporary variables used for calculating the NDVI.
Specify output path and file name (e.g.
agb_masked.tif) under Output raster layer to save the result in your working directory.Click run to start the calculation. A new raster dataset
agb_masked.tifwill appear in the Data Sources panel. The resulting raster now displays AGB estimates for forest areas, while any other land cover types are masked.
2.4.5. 5. Apply the forest mask to the NDVI
Apply the forest mask to the NDVI by using Raster Math.
mask = forest_mask.astype(bool)
ndvi_masked = ndvi
ndvi_masked[mask] = -99
ndvi_masked.setNoDataValue(-99)
del mask
Specify output path and file name (e.g.
ndvi_masked.tif) under Output raster layer to save the result in your working directory.Click run to start the calculation. A new raster dataset
ndvi_masked.tifwill appear in the Data Sources panel. The resulting raster now displays the NDVI for forest areas, while any other land cover types are masked.
2.4.6. 6. Visualize AGB vs. NDVI with the Scatter Plot tool
Close all Map Views.
Open an RGB composite of
enmap_sonoma.tifin Map #1.Display
agb_masked.tifandndvi_masked.tifin Map#2 and Map#3, respectively and use the Layer Properties to change the color ramp of both maps to white-green. Link all Map Views.
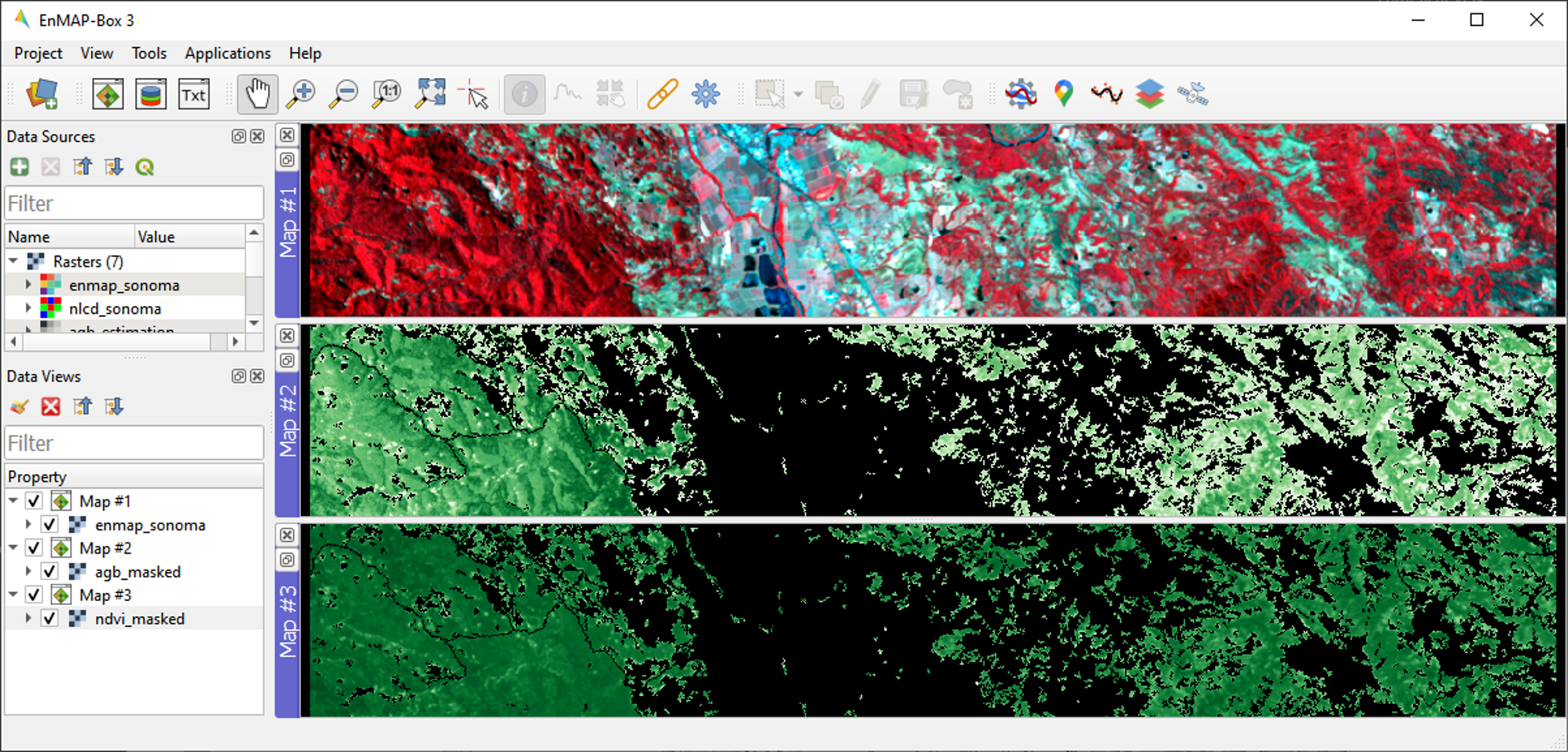
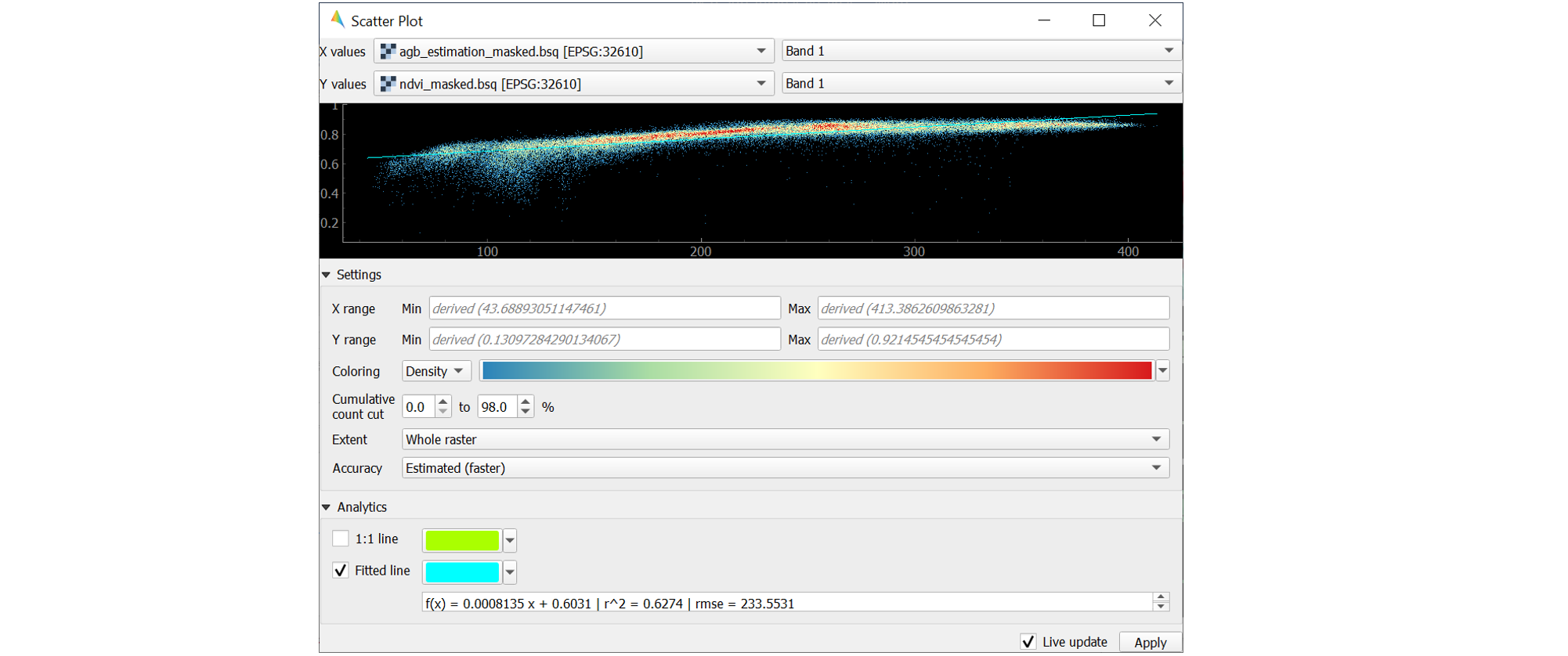
To investigate the relationship between estimated AGB and NDVI, we will make use of the EnMAP-Box’s Scatter Plot tool. This is one of several tools integrated into the EnMAP-Box to support data visualization and assessment
Open the Scatter Plot tool by going to Tools then selecting Scatter Plot.
Select
agb_masked.tifand Band 1 for specifying X values andndvi_masked.tifand Band 1 for specifying Y values. If a selected raster has multiple bands, you would specify the desired band from the dropdown.Click on Apply to visualize the Scatter Plot.
- The
Settings offer different options for visualizing the scatter plot.
You may change the Min and Max values, Coloring or Cumulative count cut options to improve your visualization.
Under Extent you may choose
Current canvasorWhole rasterto display data of the current map canvas only or to display all raster data. ChooseWhole raster.Under Accuracy you may choose to display
EstimatedorActual.Actualwill display all available data, whileEstimatedwill only display a random subset. For large raster extents,Estimatedis much faster, and for that reason is the default. Leave Accuracy asEstimated.Click on Apply to update the Scatter Plot. If Live update is checked, visualization of the scatter plot will be automatically updated.
- The
The
Analytics offers options to assess the relationship between x and y values.
The 1:1 line represents the linear 1:1 relationship between the two variables of the same unit if they were perfectly correlated.
The Fitted line represents the linear regression line fitted between the actual data from the two variables. The linear regression function, the coefficient of determination (r^2) and the Root Mean Squared Error (rmse) will be additionally displayed. Activate the Min-max line by checking the box to its left.
2.4.7. Learning Activities
Learning Activities
C1: Why was it necessary to mask the AGB results?
C2: What relationships can you see between AGB and NDVI? Do these relationships hold true if you look at the un-masked AGB and NDVI maps?
C3: Given the relationships between AGB and NDVI, do you think NDVI could be used to map AGB? What limitations would you expect from such a model?
2.5. Additional Exercises
Learning Activities
AE1: Use the Image Statistics tool in the Tools menu to look at the band statistics for the biomass predictions both with and without the tree mask applied.
AE2: Because we randomly subsetted the training data prior to model training, the performance of the model has an element of uncertainty to it. To better understand this, rerun the regression workflow 3-5 times. Then use the Raster Math tool to calculate the average estimate and variance. How does running the regression in such an ensemble approach affect the results? What is the spatial pattern of variation in estimates?
AE3: Rerun regression (Exercise B) using NDVI as the input rather than the hyperspectral imagery.
