Filtering
The EnMAP Box comes with a variety of filtering algorithms. You can find them in the processing toolbox under . In this section we will demonstrate how to apply them, by showing a spatial filter and a spectral filter.
Spatial Filter
Spatial filters apply a 2D kernel spatially (i.e. y/x dimensions) for each band.
Open the testdataset. In the processing toolbox go to .
Select
enmap_potsdam.bsqas RasterUse the default settings in the code window
Specify Output Raster, and click Run
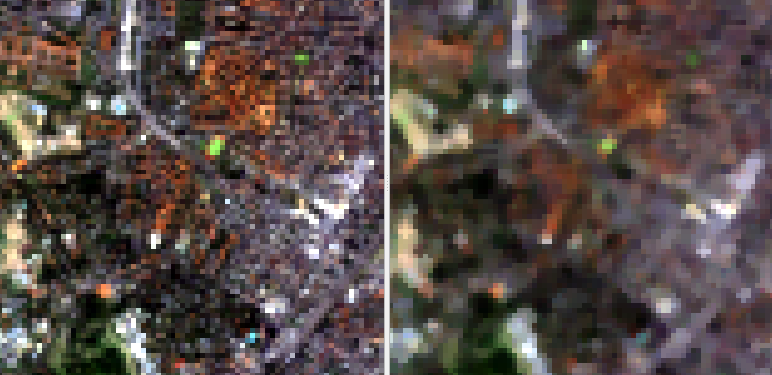
Fig. 9 Input image on the left (true color RGB), median filtered image on the right (same band combination)
Spectral Filter
In spectral filtering a 1D kernel is applied along the z dimension of the raster (bands). Mind that in some cases you will reduce the overall number of band values as values in the beginning and at the end will be set to nodata (depending on the kernel type and size).
Open the testdataset. In the processing toolbox go to .
Select
enmap_potsdam.bsqas RasterIn the code window, change
stddev=1tostddev=2Specify Output Raster, and click Run
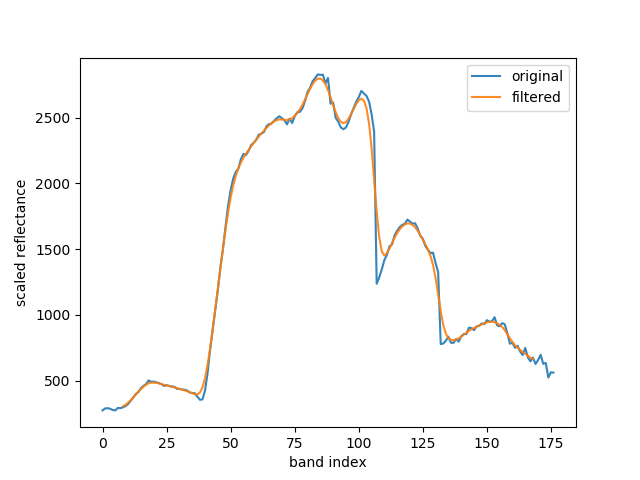
Fig. 10 Spectral signature of a pixel, original vs filtered with a Gaussian1DKernel